Multiomic cytometry
Highly sensitive detection of 5’ mRNA with antigen specificity and phenotyping.
Complete immune repertoire
Paired, full-length Ig sequence, including isotypes, and T-cell receptor α/β.
Immune mapping
Combine immune repertoire with antigen specificity at single cell resolution.
Streamlined data analysis
Explore and visualize clonotype information with easy-to-use software.
One-day lab workflow
From sample to sequencing-ready library in one day, with an option for an automated workflow.
Scalable solutions
From standard experimental needs to higher throughput for comprehensive studies, find the kit that fits your research.
Explore what you can do
Discover novel cell types and functions
Recover a comprehensive immune repertoire
Explore immune receptor mapping
Discover new biomarkers
Flexible solutions to meet your needs
Single cell resolution for comprehensive immune profiling
Multiomic analysis
Simultaneously measure cell surface protein expression and antigen specificity with 5’ gene expression.
Immune Repertoire Profiling
Understand clonal expansion and diversity by capturing full-length, paired B-cell and T-cell receptors at a single cell level.
- Paired receptor sequence enables re-expression and testing for antigen binding and function
- Combined with gene expression data, perform immune cell classification and discover new cell states
Cell Surface Protein
Correlate gene expression measurements with immunophenotyping at a single cell level to better delineate immune cell subsets and functional states.
- Enable ultra-high parameter discovery, using multiomic cytometry to detect up to hundreds of proteins simultaneously with unbiased gene expression profiling
- Stratify immune cell populations by correlating protein isoforms with regulatory network co-expression in single cells
Antigen Specificity
Assess the specificity of the adaptive immune system at single cell resolution with simultaneous capture of paired T-cell receptors and peptide–MHC multimers, or immunoglobulin heavy and light chains and their cognate antigens.
- Explore public and private T-cell antigen specificities using barcoded peptide–MHC multimers
- Characterize how neoantigens may be targeted by the adaptive immune system
- Accelerate antibody discovery with a faster and higher throughput screening technology
Large-scale projects
Tackle your most ambitious projects, from rare cell type discovery to translational studies, with our high-throughput (HT) kit and complementary solutions for larger-scale projects.
Single Cell Immune Profiling HT
Transform single cell immune profiling with high-throughput (HT) multiomic analysis on the new Chromium X, and reveal the incredible diversity of the adaptive immune system.
- Accelerate time to results
Analyze hundreds of thousands of single cells in a single chip for up to 16 samples at once.
- Enjoy economy of scale
Take advantage of high-throughput capabilities and reduce experimental and analysis costs even further with Targeted Gene Expression.
- Support from prep to analysis
From technical resources to user-friendly, cloud-based data analytics, you are supported end-to-end.
Targeted Gene Expression
Profile a defined set of transcripts to reduce costs while increasing sample number or sequencing depth with customizable, comprehensive gene panels.
- Built to scale
Maximize sample throughput and reduce hands-on time by multiplexing libraries during target enrichment.
- Seamless integration
Don’t miss any data—seamlessly transition between whole transcriptome and targeted analysis from the same library.
- More for less
As studies scale, maximize your budget by decreasing sequencing costs with pre-designed or custom targeted panels.
Workflow automation
Maximize lab productivity and obtain consistent, reproducible results with Single Cell Immune Profiling for Chromium Connect.
Automated Single Cell Immune Profiling
Automate your immune profiling workflow on the Chromium Connect to explore T- and B-cell repertoire diversity, identify rare cell populations, and reveal clonal expansion in response to disease and therapy with increased productivity.
- Optimize productivity
Shorten your hands-on time, and go from single cells to sequencing-ready immune profiling libraries with walk-away convenience.
- Standardize your workflow
Generate consistent and reproducible single cell immune profiling results across experiments, users, and even across multiple sites.
- Streamline operations
Utilize pre-aliquoted, automation-specific reagents that are color-coded and 2D-barcoded for error-proof reagent tracking and loading.
Proven results
Proven results
Publications
Search hundreds peer-reviewed publications using Chromium Single Cell Immune Profiling.
High-Throughput Mapping of B Cell Receptor Sequences to Antigen Specificity
High-Throughput Mapping of B Cell Receptor Sequences to Antigen Specificity
Cell. 2019, Ian Setliff, et al.
Cell. 2019, Ian Setliff, et al.
Massively Parallel Single-Cell B-Cell Receptor Sequencing Enables Rapid Discovery of Diverse Antigen-Reactive Antibodies
Massively Parallel Single-Cell B-Cell Receptor Sequencing Enables Rapid Discovery of Diverse Antigen-Reactive Antibodies
Communications Biology. 2019, Leonard D. Goldstein, et al.
Communications Biology. 2019, Leonard D. Goldstein, et al.
Clonal replacement of tumor-specific T cells following PD-1 blockade
Clonal replacement of tumor-specific T cells following PD-1 blockade
Nature Medicine. 2019, Katheryn E. Yost, et al.
Nature Medicine. 2019, Katheryn E. Yost, et al.
A cell atlas of human thymic development defines T cell repertoire formation
A cell atlas of human thymic development defines T cell repertoire formation
Science. 2020, Jong-Eun Park, et al.
Science. 2020, Jong-Eun Park, et al.
Resources
Find the latest app notes and other documentation Chromium Single Cell Immune Profiling.
A New Way of Exploring Immunity—Linking Highly Multiplexed Antigen Recognition to Immune Repertoire and Phenotype
A New Way of Exploring Immunity—Linking Highly Multiplexed Antigen Recognition to Immune Repertoire and Phenotype
Application Note, 10x Genomics
Application Note, 10x Genomics
Improving Single Cell Characterization with Simultaneous Gene Expression and Cell Surface Protein Measurements at Scale
Improving Single Cell Characterization with Simultaneous Gene Expression and Cell Surface Protein Measurements at Scale
Application Note, 10x Genomics
Application Note, 10x Genomics
Characterization of Tumor Microenvironment
Characterization of Tumor Microenvironment
Application Note, 10x Genomics
Application Note, 10x Genomics
Our end-to-end solution

Chromium instruments with Next GEM technology
Our scalable instrument
Our compact instrument
Our automated instrument

Chromium Single Cell Immune Profiling reagents
With our reagent kits, explore adaptive immunity through multidimensional approaches, including 5’ mRNA, full-length paired BCR and TCR sequences, antigen-receptor specificity, cell surface proteins, and more.
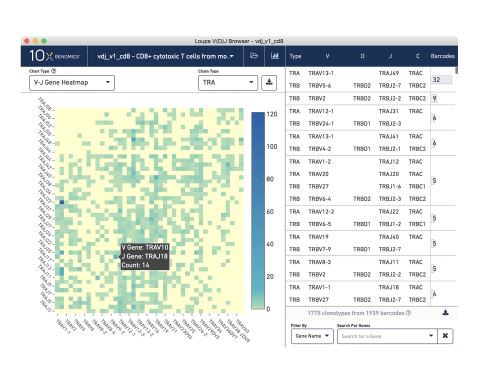
Analysis and visualization software
Our analysis pipelines
Our visualization software

World-class technical and customer support
Our expert support team can be contacted by phone or email.
Workflow
The base workflow for Single Cell Immune Profiling is shown below. If you’re using our Feature Barcoding Kit for Cell Surface Protein or Antigen Specificity, we provide additional protocols for modifying your cells of interest. For the automation option, separate reagents and protocols are available for Chromium Connect.
- 1
Prepare your sample
Start with hundreds to hundreds of thousands of cells. Cells can be labeled with antibodies and/or peptide–MHCs conjugated with a unique oligonucleotide barcode. You can use cell suspensions from FACS/MACS, fine needle aspirates, TILs, PBMCs, and more.
Resources - 2
Construct your 10x library
Construct a 10x barcoded library using our reagent kits and a compatible Chromium instrument. Each member of the Chromium instrument family encapsulates each cell with a 10x barcoded Gel Bead in a single partition. Within each nanoliter-scale partition, cells undergo reverse transcription to generate cDNA, which shares a 10x Barcode with all cDNA from its individual cell of origin.
Resources- User Guide / Protocol
HT User Guide / Protocol - Coming Soon
- Automated User Guide / Protocol
- 3
Sequence
The resulting 10x barcoded library is compatible with standard NGS short-read sequencing on Illumina sequencers, for immune and transcriptional profiling of thousands of individual cells.
Resources - 4
Analyze your data
Convert raw sequencing data into biologically meaningful insights with Cell Ranger. Cell Ranger processes data from paired immune receptor repertoire sequences, gene expression, and antigen specificity.
Analysis pipelines output
Output includes QC information and files that can be easily used for further analysis in our Loupe Browser visualization software or third-party R or Python tools.
Resources - 5
Visualize your data
Use our Loupe Browser visualization software to interactively explore your results and compare detailed clonotype information within and across samples and cell types to study gene usage, repertoire diversity, clonal expansion, and more.
Do I need to be a bioinformatician to use it?
Loupe is a point-and-click software that’s easy for anyone to download and use.
Resources
Frequently asked questions
The solution is capable of the modular integration of multiple data types, including mRNA-based whole transcriptome or targeted gene expression, immunophenotyping, adaptive immune repertoire clonality and diversity, and antigen specificity, simultaneously from the same single cells. This multiomic approach to cell characterization helps decipher the rules of the immune system one cell at a time.
Yes. You can amplify only the immunoglobulin or TCR libraries for sequencing and save the amplified whole transcriptome cDNA material for later sequencing.
Chromium Single Cell Immune Profiling is compatible with fresh or cryopreserved single cell suspensions. Typical samples include PBMCs, tumor-infiltrating lymphocytes, spleen, bone marrow cells, and other tissue types. Species can range from human, mouse, rat, and other model organisms. However, only human and mouse are currently supported by 10x Genomics reagents and protocols for immunoglobulin or TCR sequencing.
The Chromium Single Cell Immune Profiling Solution makes use of gene-specific amplification primers that target the constant regions of Ig and TCR genes in order to enrich and sequence across the entire Variable, Diversity, and Joining gene segments.
Yes, immunoglobulin heavy chain sequences are classified by IgA, IgD, IgE, IgM, and IgG.