Resources:
Scalable antigen screening
Antigen screening of up to hundreds of thousands of T and B cells in a single reaction.
Actionable insights
In days, get functional immune receptor sequences alongside antigen specificity.
Flexible sample input
Compatible with magnetic bead–based and flow sorting sample enrichment methods.
Compatible partners
Leverage the expertise of our compatible partners for off-the-shelf or custom products.
Single integrated workflow
From cells to antigen-specific receptor sequences for discovery of novel TCRs or antibodies.
Streamlined data analysis
Explore and visualize clonotypes, repertoire diversity, clonal expansion, and more.
Explore what you can do
B-cell receptor mapping at scale
Gain an understanding of your T-cell repertoire specificity
Comprehensive characterization of the immune response to antigens
Scale your investigation of antigen specificities with ease
Proven results
Proven results
Publications
Search hundreds of peer-reviewed Immunology publications using 10x Genomics products.
High-Throughput Mapping of B Cell Receptor Sequences to Antigen Specificity
High-Throughput Mapping of B Cell Receptor Sequences to Antigen Specificity
Cell, 2019, Ian Setliff et al.
Cell, 2019, Ian Setliff et al.
Massively Parallel Single-Cell B-Cell Receptor Sequencing Enables Rapid Discovery of Diverse Antigen-Reactive Antibodies
Massively Parallel Single-Cell B-Cell Receptor Sequencing Enables Rapid Discovery of Diverse Antigen-Reactive Antibodies
Communications Biology, 2019, Leonard D. Goldstein et al.
Communications Biology, 2019, Leonard D. Goldstein et al.
Human Thymopoiesis Selects Unconventional CD8+ α/β T Cells That Respond to Multiple Viruses
Human Thymopoiesis Selects Unconventional CD8+ α/β T Cells That Respond to Multiple Viruses
bioRxiv, 2020, Valentin Quiniou et al.
bioRxiv, 2020, Valentin Quiniou et al.
Resources
Find the latest app notes and other documentation.
A New Way of Exploring Immunity—Linking Highly Multiplexed Antigen Recognition to Immune Repertoire and Phenotype
A New Way of Exploring Immunity—Linking Highly Multiplexed Antigen Recognition to Immune Repertoire and Phenotype
Application Note, 10x Genomics
Application Note, 10x Genomics
Redefining Cellular Phenotyping: Comprehensive Characterization and Resolution of the Antigen-Specific T Cell Response
Redefining Cellular Phenotyping: Comprehensive Characterization and Resolution of the Antigen-Specific T Cell Response
Application Note, 10x Genomics
Application Note, 10x Genomics
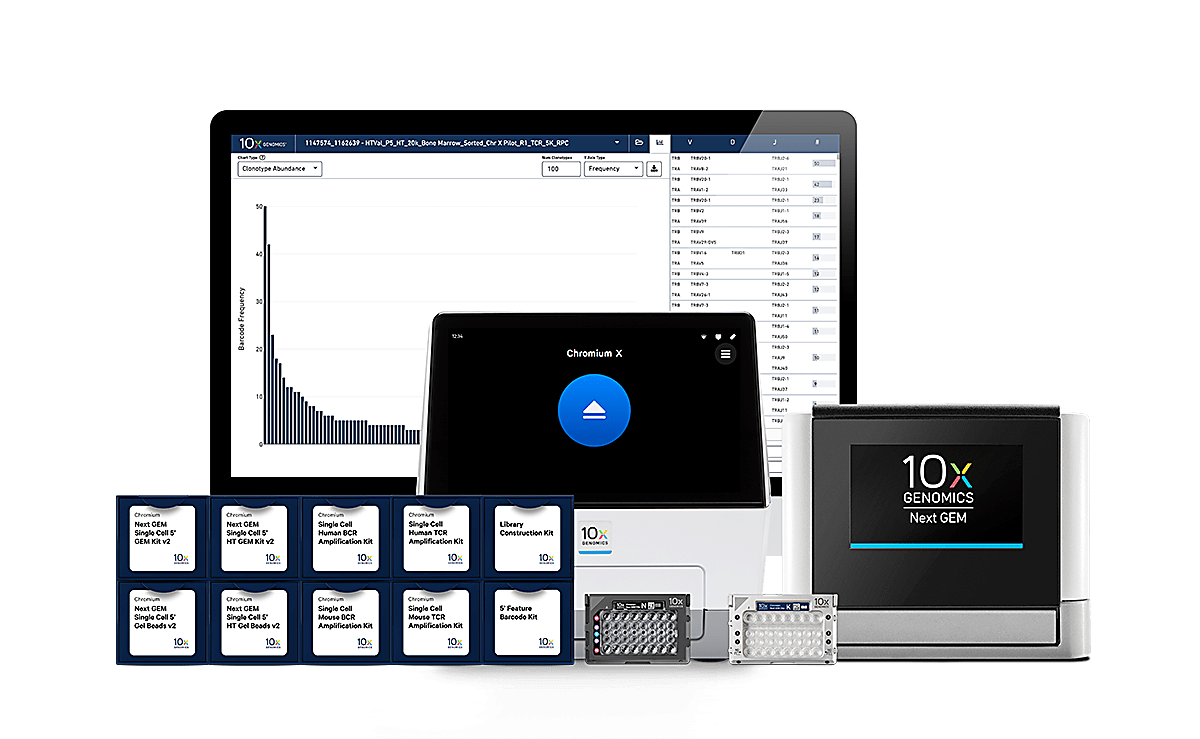
Our end-to-end solution

Chromium instrument with Next GEM technology
Our scalable instrument
Our compact instrument
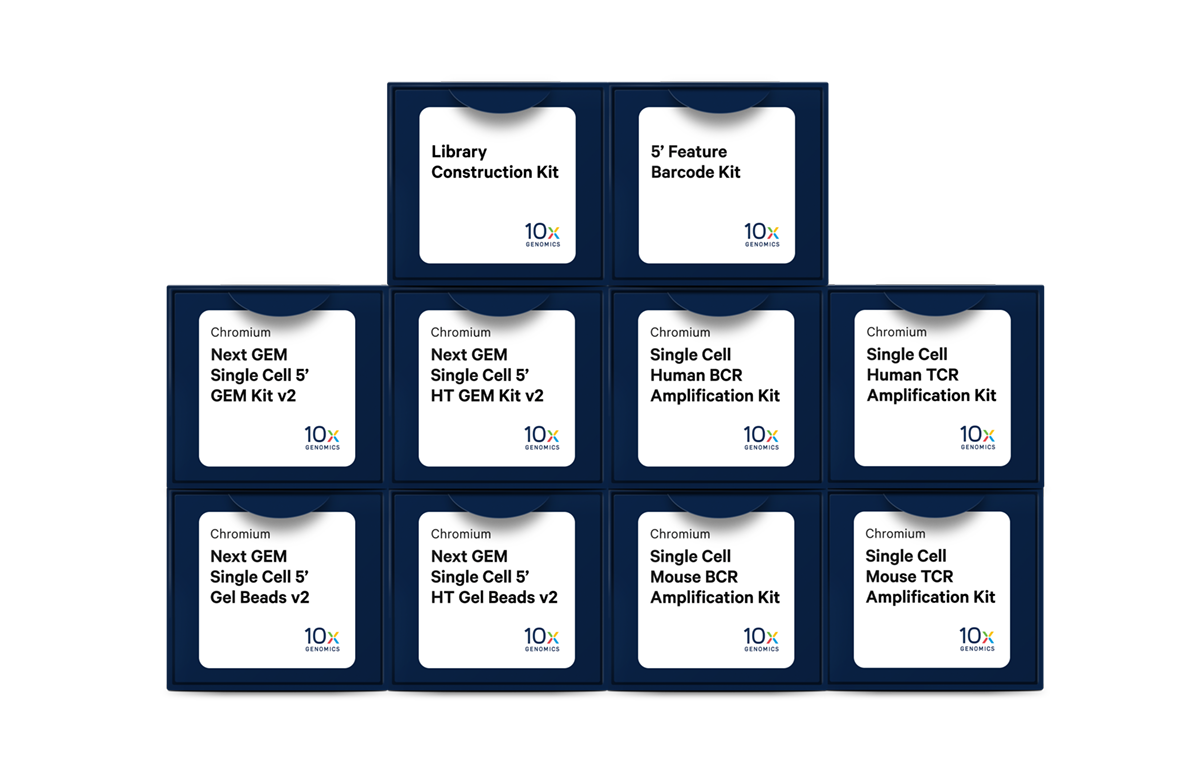
Chromium Single Cell Immune Profiling reagents
With our reagent kits, explore adaptive immunity through multidimensional approaches, including 5’ mRNA, full-length paired BCR and TCR sequences, antigen-receptor specificity, cell surface proteins, and more.
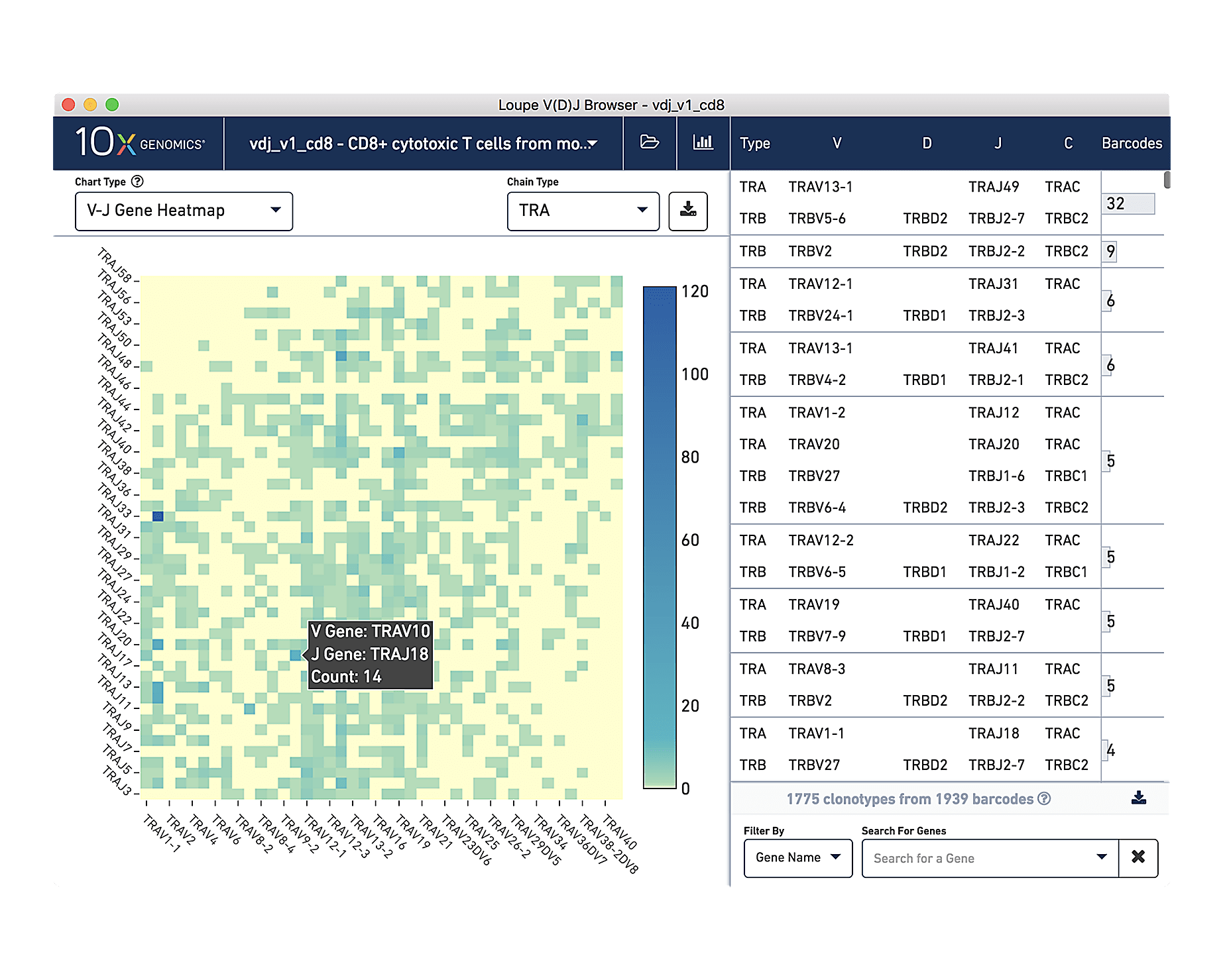
Analysis and visualization software
Our data processing platform
Our analysis pipelines
Our visualization software

World-class technical and customer support
Our expert support team can be contacted by phone or email.
Components required
Single Cell Immune Receptor Mapping is enabled with the following 10x Genomics products:
- Chromium Next GEM Single Cell 5' Kit v2
1000265, 4 rxns | 1000263, 16 rxns)
- Chromium Next GEM Single Cell 5’ HT Kit v2
(1000374, 8 rxns | 1000356, 48 rxns)
- Chromium Next GEM Chip K Single Cell Kit
(1000287, 16 rxns | 1000286, 48 rxns)
- Library Construction Kit
(1000190 | 16 rxns)
- Dual Index Kit TT Set A
(1000215 | 96 rxns) - Gene Expression and/or Immune Repertoire Profiling Libraries
- 5' Feature Barcode Kit
(1000256 | 16 rxns)
- Dual Index Kit TN Set A
(1000250 | 96 rxns) - Feature Barcode Libraries
- Chromium Single Cell Human TCR Amplification Kit
(1000252 | 16 rxns)
- Chromium Single Cell Human BCR Amplification Kit
(1000253 | 16 rxns)
- Chromium Single Cell Mouse TCR Amplification Kit
(1000254 | 16 rxns)
- Chromium Single Cell Mouse BCR Amplification Kit
(1000255 | 16 rxns)
- Targeted Gene Expression
BioLegend TotalSeq™ antibodies
Workflow
- 1
Prepare your sample
Label your cells with desired antigens or pMHC multimers conjugated with a unique oligonucleotide barcode. Depending on the expected percentage of antigen-specific cells, cell populations can be enriched through flow-based or magnetic sorting. Compatible cell suspensions include samples from dissociated tumor cells, fine needle aspirates, TILs, PBMCs, and more.
Resources - 2
Construct your 10x library
Construct 10x barcoded libraries using our reagent kits and a compatible Chromium instrument. Each member of the Chromium instrument family encapsulates each cell with a 10x barcoded Gel Bead in a single partition. Within each nanoliter-scale partition, cells undergo reverse transcription to generate cDNA, which shares a 10x Barcode with all cDNA from its individual cell of origin.
Resources- User Guide / Protocol for Immune Profiling
HT User Guide / Protocol for Immune Profiling - Coming Soon
- 3
Sequence
The resulting 10x barcoded libraries are compatible with standard NGS short-read sequencing on Illumina sequencers for immune and transcriptional profiling of thousands of individual cells.
Resources - 4
Analyze your data
Convert raw sequencing data to biologically meaningful insights with Cell Ranger. Cell Ranger processes data from gene expression, paired immune receptor repertoire sequences, cell surface proteins, and antigen specificities.
Analysis pipelines output
Output includes QC information and files that can be easily used for further analysis in our Loupe Browser visualization software, or third-party R or Python tools.
Resources - 5
Visualize your data
Use our Loupe Browser and Loupe V(D)J Browser software to define and compare cell types based on different marker types.
Do I need to be a bioinformatician to use it?
Loupe is a point-and-click software that’s easy for anyone to download and use.
Resources
Frequently asked questions
10x Genomics offers a fully supported workflow for determining antigen-specificity in T cells through use of Immudex’s dCODE Dextramer® reagents. This workflow can identify paired receptors that bind your epitope of interest at the single cell level. A technique recently reported by Vanderbilt can be used to determine B-cell antigen specificity (Ian Setliff et al., High-Throughput Mapping of B Cell Receptor Sequences to Antigen Specificity. Cell, doi: 10.1016/j.cell.2019.11.003).
Antigen specificity data can be used for a wide range of assay types, including high-throughput antibody discovery, vaccine design, neo-antigen identification, high-resolution immune profiling studies, and more.
Most multimer assays are fluorescence-based, meaning that the multimer molecules are tagged with a fluorescent marker that can then be used for sorting antigen-specific cells. These kinds of studies can provide information about the number of antigen-specific cells in a cell population. With Single Cell Immune Profiling, you can get this information, as well the paired, full-length sequences of the antigen-specific receptors and gene expression data at the single cell level. Together, this data provides a complete profile of the immune response.
Because antigen-specific populations are rare, we recommend sorting for antigen-positive cells prior to 10x Genomics library preparation, but, ultimately, it depends on the goals of the experiment.
See our Demonstrated Protocol on how to label cells for antigen-specificity staining. Additional resources can be found at the Immudex website.